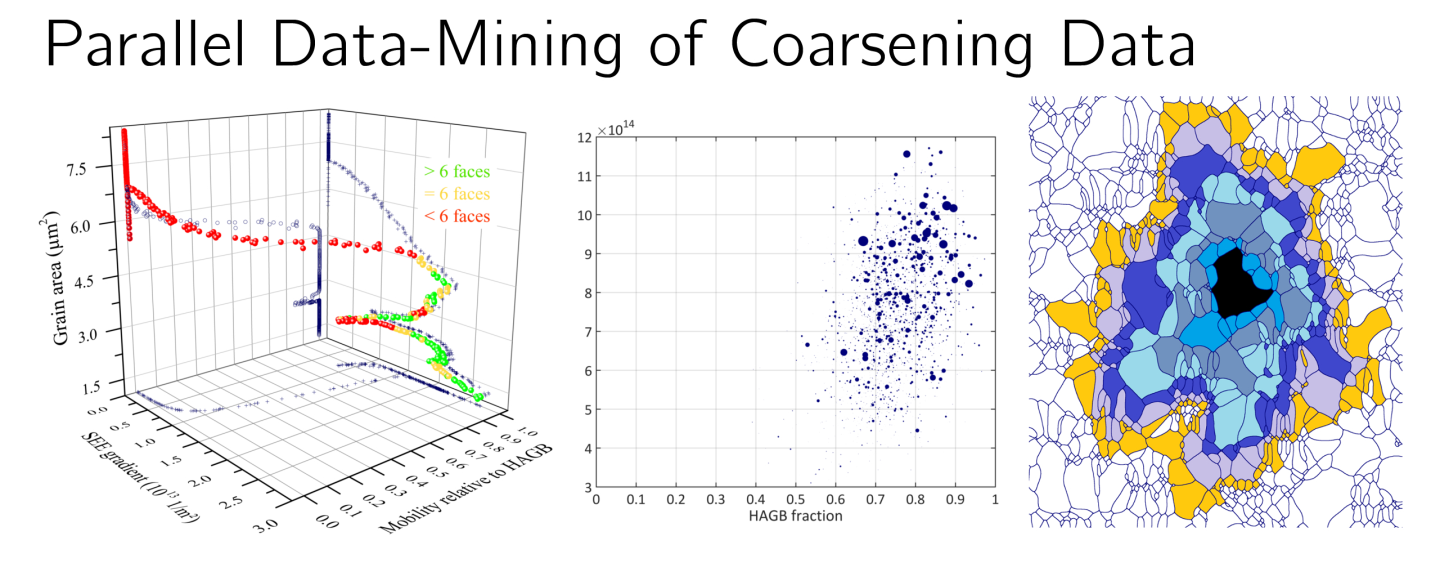
TopologyTracer is an MPI/OpenMP-parallelized datamining tool for conducting spatio-temporal analyses of microstructural element dynamics. Its purpose is the quantification of correlations — spatial and temporal — between individual microstructural elements and their higher-order neighboring elements. Currently, the software allows comprehensive studies of individual grains’ volume evolution and set their growth history into relation to the evolution of their higher-order neighbors. The tool is unique in two ways: on the one hand in its ability to process these surveys in parallel and thereby handle hitherto intractable large datasets including millions of grains. On the other hand in its capability to explicitly take the spatial distribution of the long range neighborhood into account.
The source code was developed by Markus Kühbach during his PhD time with Luis A. Barrales-Mora and Günter Gottstein at the Institute of Physical Metallurgy and Metal Physics (IMM) with RWTH Aachen University. Being now with the Max-Planck-Institut fur Eisenforschung GmbH in Düsseldorf, I maintain the code, though at disregular intervals. Nonetheless, feel free to utilize the tool, do not hesitate contacting me for sharing thoughts, suggesting improvements, or reporting your experiences.
1. Getting started¶
2. Creating input¶
3. TopologyTracing¶
TopologyTracing requires at least to have
- A collection of ID contiguous binary data file pairs, i.e. Faces_*.bin and Texture_*.bin binary files,
- The topotracer2d3d executable,
- A XML control file all in the same folder.
Several analysis routines require additional files as it is explained for many in the input section.
- With these prerequisites, TopologyTracing, i.e. the datamining of simulation data, requires two steps:
- 1. Setup the control file2. Specify the post-processing tasks
Both tasks are accomplished via the XML control file:
4. Visualizing results¶
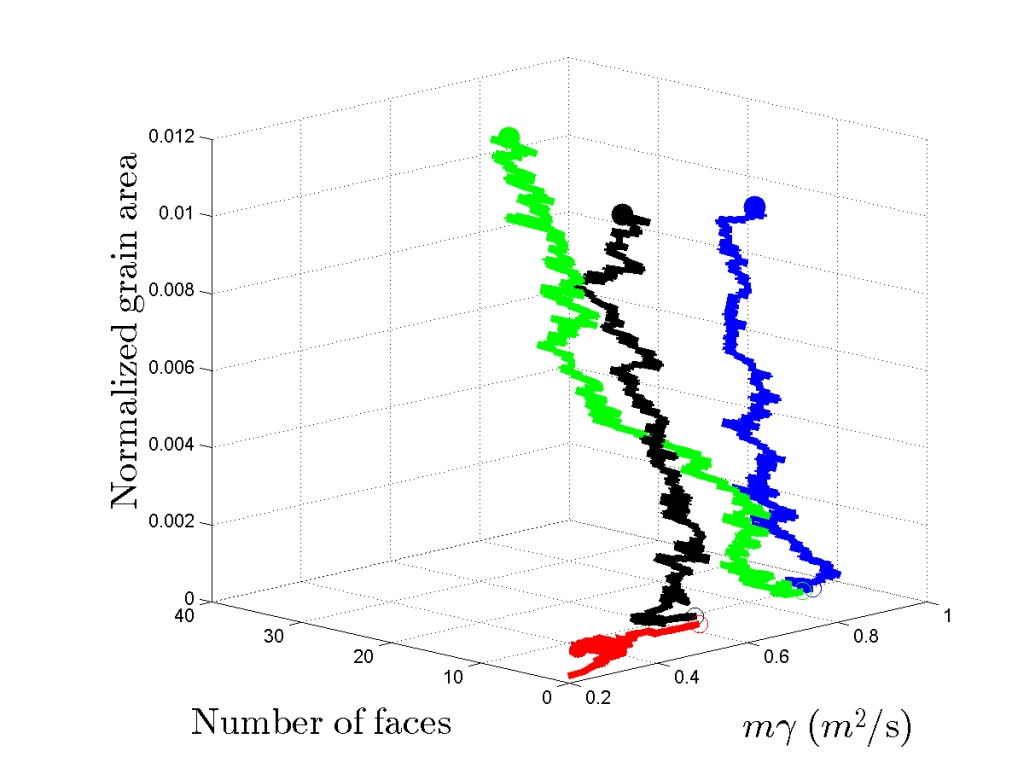
In its current state, the TopologyTracer does not contain a GUI. Instead, plain MPI I/O raw files and ASCII files are generated which require post-processing with for instance Matlab scripts available in the scripts folder. In general one can visualize a growth path by the plot3(X,Y,Z) command and access a distribution of values via the histogram function. Therein X, Y, Z specify each a row vector of length NC for a particular grain ID in row r.
Funding¶
Version history¶
Questions, contributions¶
Just let me know or contact m.kuehbach@mpie.de