In a nutshell¶
What is the TopologyTracer?¶
Currently, a software for the datamining of dump snapshots obtained from densely sampled instrumented grain coarsening simulations.
What are the user benefits?¶
What sets the TopologyTracer apart from other tools like DREAM3D or MTex?¶
While the aforementioned tools’ strength is their ability to compute distributional descriptive statistics, like the grain size distribution or the orientation distribution function (ODF), they fall still short in their spatio-temporal and correlative capabilities. Additionally, they have much lower scalability as they do so far not employ parallelization. Thus, it remains the user’s responsibility to add parallel functionality into the source code or set up batch processing queues via modifying the analysis scripts. Either way, this is not only a tedious task but in particular with respect to the scripting environment not straightforward to implement in a manner to advise the script interpreter exactly and in full control of the user were data should be stored to employ as much memory locality as possible: that is to assure that data which describe the microstructure in local regions are mapped as closely as possible into main memory such that their processing requires less costly memory queries.
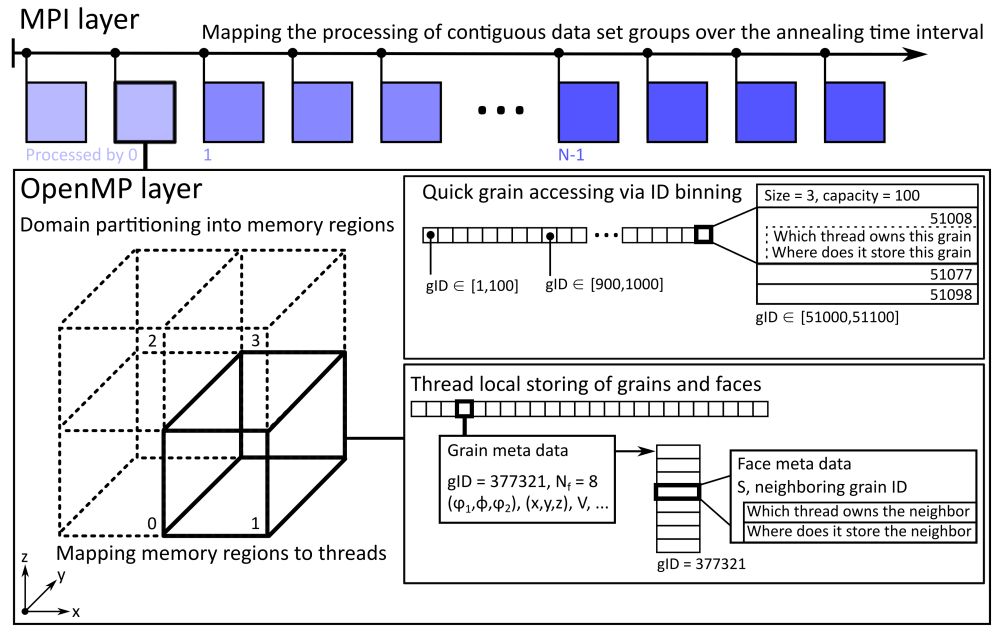
By contrast, the TopologyTracer improves on this with its two-layer data parallelism. The individual datasets, each of which represents one time step, are elements in the coarser MPI-layer. Each MPI process hosts a contiguous set of all datasets over a particular time step interval. Additionally, each dataset is stored spatially partitioned into memory regions. These become mapped in such a manner as to reduce the number of remote memory accesses. It is exactly this data-locality-improvement-oriented design which suits potential thread-parallel execution with OpenMP for instance. Several pieces of this MPI/OpenMP-hybrid parallel scheme have already been implemented.
The figure illustrates this two-level parallelism.
In particular the logical and physical partitioning of the data in the second layer is fundamental to achieve scalable performance by improving data locality. This stands in clear contrast to many proprietary Materials Science software packages which still rely on traditional data structures commonly employed during sequential programming and their execution via scripting-language-based post-processing during which the complexity of data placement is opaque to the user. This opacity is the key to making perceivingly easily maintainable most Computational Materials Science scripting workflows. At the same time, though, it is the prime brake for achieving performance and thereby productive workflows - in particular for ever growing datasets. Admittedly, one could also employ thread parallelism without worrying about locality aspects. Beyond some problem-specific thread count threshold, though, this practice results in increasingly less performant execution as internally frequent remote memory accesses will be required. Exactly, these will then effectively saturate most contemporary memory infrastructures on not only super computers but also workstations and therefore prevent the quick delivering of data to the floating point processing units.